Tumors are complex ecosystems where in addition to the malignant cells, multiple stromal and immune (including lymphoid and myeloid subsets) populations also coexist. The recent clinical successes of immunotherapies targeting checkpoint inhibitors such as PD-1, have shed light on the crucial role played by tumor infiltrating lymphocytes (TILs) and has led new clinical successes. However, as not all patients respond to immunotherapies, there is a crucial need to find new targets and even to combine them to cure non-responders. Innate immune cells are highly represented in tumors, including tumor-associated macrophages (TAMs) being one of the major immune populations.
Macrophages have emerged as immune cells with a broad spectrum of non-immune related tissue-supporting activities and they play important roles in the tumour microenvironment (TME), as an important component of the tumor microenvironment and the concordance between experimental and clinical data towards a pro-tumoral effect of TAM. Moreover, TAMs not only express PD-L1, but also PD-1 and are therefore targeted in current immunotherapy protocols using anti-PD-1. However, the clinical implications of their targeting remain mostly unknown, and our knowledge on their functions within the TME is still sparse. It is essential to elucidate the negative implications of TAMs in the TME as they represent pertinent cellular targets to design novel immunotherapies. Currently there are already several trials targeting the colony stimulating factor 1 receptor (CSF-1R) or CSF-1. Other clinical trials evaluating TAM-targeted drugs are already on-going, but the majority of them being still in phase I/II. These clinical trials are following two main strategies: (i) reprograming pro-tumoral TAM to an anti-tumoral phenotype and (ii) reducing TAMs infiltration, either by inhibiting TAM-recruitment or by directly killing them. So far, clinical translation of these strategies is limited. The most concerning is that current strategies do not address TAMs diversity according to tumor types, since we still lack such knowledge. Unfortunately, these approaches do not take into consideration the heterogeneous populations of TAM, but only approaching them as a single population to target.
Tissue-resident macrophage (TRM) identity and functions are shaped by various intrinsic and extrinsic factors among which ontogeny, i.e., developmental origin, has a prominent role. In recently years our laboratory pioneered, and established, that macrophages could arise from three different waves occurring sequentially during development. Adult TRMs comprise a defined mixture of macrophages that arise from either embryonic cell or adult monocytes. Each tissue appears to be a mosaic composed by these ontogenically distinct populations in a tissue-specific relative abundance. Moreover, TRM identity is constantly shaped by local factors unique to each niche of residence, evolving across development, during ageing, and through inflammation. On top of these features is superimposed the cumulative effect of time spent in a specific tissue, which contributes to the acquisition of a unique signature that represents a resilient adaptation to their dynamic environment. Transposing these concepts to cancer, we hypothesize that the TME is likely to contain a mixture of TAMs that have spent different amounts of time within the tissue and in contact with tumor cells; ranging from monocyte-derived macrophages that were freshly recruited and differentiated a few hours/days ago, to those that have resided there for several weeks/months before or after tumor emergence, alongside original embryonic-derived macrophages that have been present since before birth. Since ontogeny and duration of residence within tissues impact the degree of differentiation and thus the transcriptional program of macrophages, considering TAMs as one population is far too simple, and naive. In addition, the precise location within the tumor of TAMs populations has never been addressed.
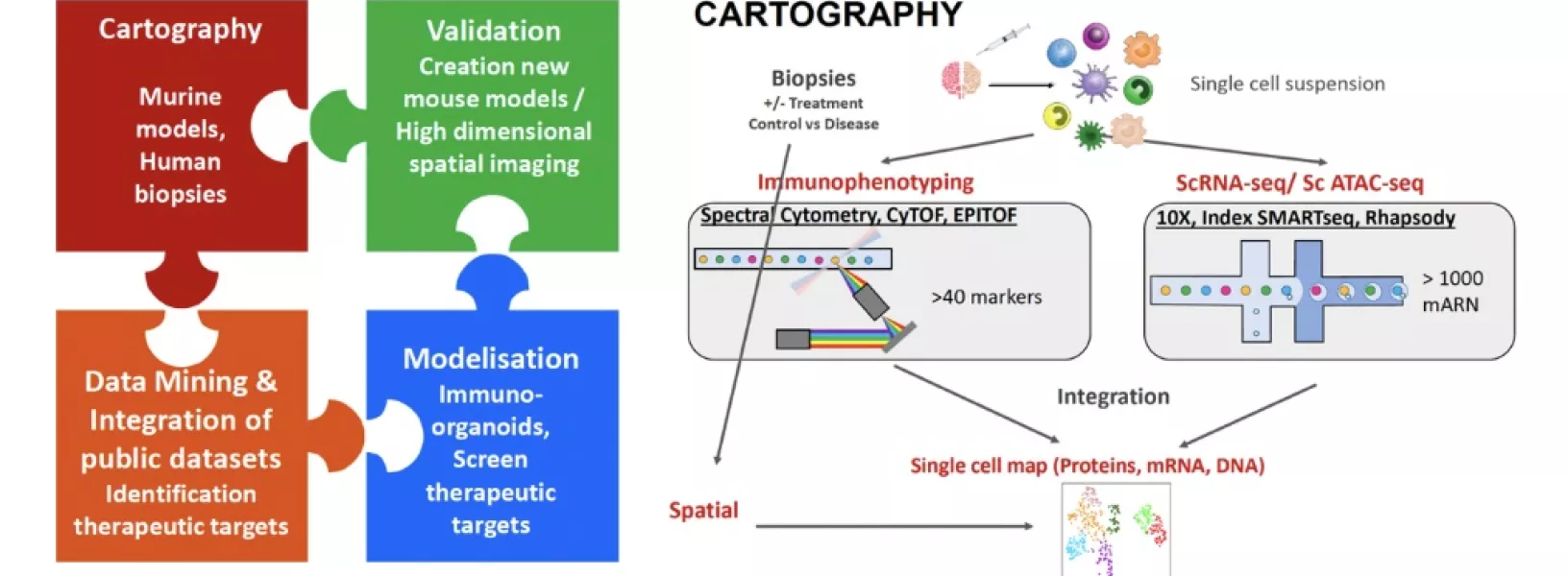
We will address myeloid heterogeneity and functions in cancer using a combination of synergistic high dimensional approaches as well as organoid/tumoroid approaches as described below:
- Cartography of myeloid cells in murine cancer models in unique proprietary mouse lines tracking macrophage heterogeneity (based on ontogeny and time) in absence or presence of relevant interventions.
- Deep cartography and referencing of published human data and unique “dynamic” cancer cohorts using integration of mRNA, protein expression, epigenetic profiling and spatial location. This will allow us to identify new myeloid targets to design new therapeutical strategies to cure cancer.
Modeling macrophage function in pediatric cancers with patient-derived 3D organoids/tumoroids (patient avatar) using induced pluripotent stem cell (IPSC)-derived macrophages.